 Viewing the SURFNET surfaces
Viewing the SURFNET surfaces
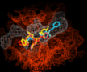 Viewing the maps in QUANTA
Viewing the maps in QUANTA
The brick maps generated for
QUANTA will have a .mbk extension.
They can be viewed in QUANTA using the menu options:-
Draw
Create Objects
Brick Maps
Enter a name for the map and press OK
Select Brick Map File and press OPEN
Pick the option All of the map displayed
Enter number of contour levels as 1
Enter contour level as 100 and select colour (1-14)
Use a single contour level of 100.0 for van der Waals surfaces and
gap regions. For density distributions, use whichever contour level is most
appropriate.
Displaying binding-site shells
You can use the QUANTA options to display binding-site masks as
shells, rather than as completely closed surfaces, as follows:-
- Firstly, select just the atoms of the protein, using:-
You can select the part of the protein you are interested in by, say, using
the Pick Segment option.
Then, select Finish.
- Set the brick-map radius to its minimum value (0.5), using:-
- Then, when loading the brick-map representing the binding-site mask,
use the Map covers displayed atoms option after the usual steps:-
Use the same contour levels as above (ie 100.0 for van der
Waals surfaces), and the map will be as before, but the contour surface
will be open wherever the gap region extends away from the protein into free
space.
 Viewing the SURFNET surfaces
Viewing the SURFNET surfaces
 Viewing the SURFNET surfaces
Viewing the SURFNET surfaces Viewing the SURFNET surfaces
Viewing the SURFNET surfaces Viewing the maps in QUANTA
Viewing the maps in QUANTA Viewing the SURFNET surfaces
Viewing the SURFNET surfaces