
 Viewing the SURFNET surfaces
Viewing the SURFNET surfaces

 Viewing the SURFNET surfaces
Viewing the SURFNET surfaces
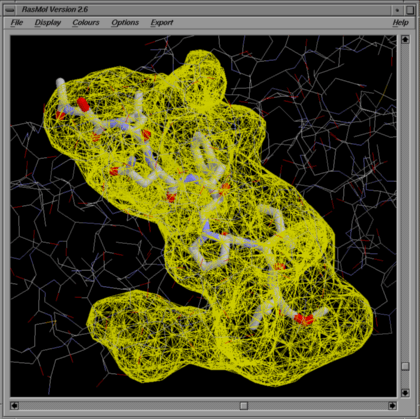
This file can be loaded into Rasmol like any other PDB file. If the data corresponds to a surface, or gap regions, of a given protein structure, you might want to view the protein together with the surfaces. As Rasmol allows only the input of a single PDB file, you can view both at the same time by concatenating surfnet.pdb with your original PDB file.
To view in Rasmol:-
RasMol> wireframe off
RasMol> select not map
RasMol> wireframe
This displays just the protein in a wireframe representation. Alternatively, you can use one of the other display options for the protein (eg backbone, ball-and-stick, ribbon, etc)
RasMol> select map
RasMol> wireframe
RasMol> colour bonds red
If more than one surface was written out to the surfnet.pdb file, you can define the one you're after by, say, select map2 to pick the second surface. This allows you to colour each surface in a different colour using the colour bonds command.
RasMol> select map
RasMol> spacefill 0.2
RasMol> colour atoms blue
As above, if more than one surface was written out to the surfnet.pdb file, you can define the one you're after by, say, select map2 to pick the second surface. This allows you to colour each surface in a different colour using the colour atoms command.

 Viewing the SURFNET surfaces
Viewing the SURFNET surfaces